#nucleosome
Text

The Nucleosome: DNA's Fancy Packaging and Party Trick!
Imagine cramming two meters of yarn into a pea-sized box. Sounds impossible, right? Well, that's the impressive feat that cells pull off every single day with DNA! They use a clever structure called the nucleosome to pack this massive genetic blueprint into the tiny nucleus.
The journey began in 1974 when Don and Ada Olins, peering through an electron microscope, spotted repeating beads – the first glimpse of nucleosomes. Roger Kornberg, building upon this observation, proposed the now-iconic "subunit theory," envisioning DNA wrapped around histone protein cores. This theory, later solidified by Pierre Oudet's term "nucleosome," laid the groundwork for further exploration. The 1980s witnessed a flurry of activity, with Aaron Klug's group using X-ray crystallography to reveal the left-handed superhelical twist of DNA around the histone octamer. But the true masterpiece arrived in 1997 when the Richmond group, armed with advanced techniques, unveiled the first near-atomic resolution crystal structure of the nucleosome. This intricate map, showcasing the precise interactions between DNA and histones, remains a cornerstone of our understanding.
The Players:
DNA: The star of the show, carrying our genetic code in the form of a double helix.
Histones: Protein spools around which DNA tightly winds. Imagine eight of them forming a core, like a mini-protein drum set.
Linker DNA: Short stretches of DNA connecting the spools, like the spaces between beads on a necklace.
The Steps:
Wrap and Roll: Picture DNA gracefully wrapping around the histone core, like thread around a spool. Each nucleosome holds about 146 base pairs of DNA, making about 1.67 turns.
Connect and Repeat: Linker DNA bridges the gap between nucleosomes, forming a "beads-on-a-string" structure. Think of it as pearls strung between the spools.
Compact and Condense: This repetitive unit folds further, creating intricate 30-nanometer fibers. Imagine these as twisted strands of pearls!
Here's the coolest part: histones aren't static. They can be chemically modified, like adding or removing phosphate groups. These modifications act like tiny flags that tell the cell how tightly to wrap the DNA, essentially throwing a "party" for specific genes by making them more accessible. This fine-tuning allows cells to respond to their environment and express the right genes at the right time. Understanding the nucleosome model is crucial for unraveling the mysteries of gene regulation and diseases like cancer. By studying how modifications affect nucleosome structure and gene access, scientists can develop new therapies to target specific genes and potentially treat diseases at the root cause.
While the nucleosome model is the foundation, the story gets even more intriguing. Different histone types and modifications create variations, influencing chromatin structure and function. Think of it as different music genres influencing the dance moves! Additionally, other proteins interact with the nucleosome, adding another layer of complexity to this fascinating choreography.
The nucleosome model is more than just a neat way to package DNA. It's a testament to the intricate dance between molecules that orchestrates life's processes. By understanding this fundamental structure, we gain deeper insights into cellular function, paving the way for advancements in medicine and beyond.
Remember, this is just the beginning! The world of nucleosomes and chromatin is vast and ever-evolving. So, keep exploring, keep questioning, and keep dancing to the rhythm of DNA!
#molecular biology#biology#science sculpt#life science#science#dna#biotechnology#genetics#Histone#Nucleosome#chromatin
8 notes
·
View notes
Text
The fundamental structural unit of chromatin is the nucleosome, an assembly consisting of a group of certain proteins, called histones (designated H1, H2A, H2B, H3 and H4, see figure 25.13a), wrapped in DNA (figure 25.13b). (...) In a nucleosome, B-DNA is wound around the histone unit by about 1.8 coils (figure 25.13b,c). (...) The nucleosomes are further folded to form a filament, with a diameter of ~30 nm, which has been proposed to have the structure down in figure 25.13d.
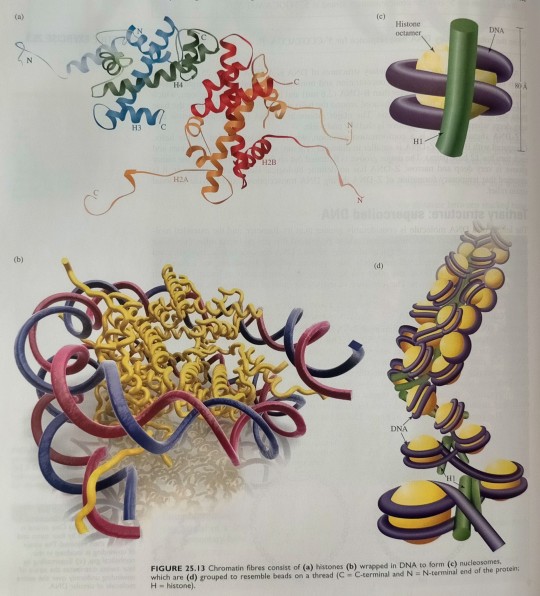
"Chemistry" 2e - Blackman, A., Bottle, S., Schmid, S., Mocerino, M., Wille, U.
#book quotes#chemistry#nonfiction#textbook#dna#chromatin#nucleosome#protein#deoxyribonucleic acid#bdna#histone#filament#folding
1 note
·
View note
Photo

#ribosomes#single#mRNA#multiple#copies#polypeptide#plastidome#polyhedral#bodies#polysome#nucleosome#science#solutions
1 note
·
View note
Text
Chemistry Notes (Sept. 2023)
Full Notes:
Henderson-Hasselbalch Equation Ex. 4
Proposed Lewis Structure Ex. 5
Red Blood Cell Disorders
Solution Equations
Steroids
.
Note Cards:
Amphipathic Compounds
Characteristics of Helices
Dynamics of Enzyme-Catalyzed Reactions
Essential Amino Acids Mnemonic
Structure of Nucleosomes
.
Patreon
#studyblr#notes#masterlist#studyblr masterlist#studyblr resource masterlist#resource masterlist#resources#studying resources#studyblr resources#sources#chemistry#chem#biochem#biochemistry#organic chemistry#orgo chem#orgo#ochem#organic chemistry resources#chemistry resources#studying chemistry#studying mcat#mcat#mcat resources#chemistry resource masterlist#master list#studyblr master list#resources master list#biochem notes#biochemistry notes
46 notes
·
View notes
Note
Hello! I am a molecular biologist, and I was wondering if I could get your opinion on some of my theories on Gallifreyans.
I haven't read through everything on your blog yet, but I'm working my way through it (lol). So some of this may not be quite accurate with what you have set up thus far.
Basically, I want to briefly discuss alternative splicing! Anyway, in metazoans, alternative splicing outcomes can be regulated in a time and tissue specific manner by legitimately hundreds of biomolecules such as RNA binding proteins, chromatin remodelers, hormones, etc etc. It is subject to epigenetic regulation as alternative splicing and transcription are coupled (and splicing largely occurs cotranscriptionally), so details such as DNA methylation, nucleosome positioning, histone modifications, etc can change the balance of different mRNA isoforms. This is largely because these factors will either help recruit splicing factors (or inhibit their recruitment) or because it will slow RNA Polymerase II elongation.
Onto my theories. I have been thinking for a little while that the lindos hormone can perhaps modulate splicing, triggering the production of regeneration-specific isoforms. Perhaps their bodies work so fast that isoforms promoting totipotency trigger a temporary transition away from the cells' differentiated states.
I also think it could be possible that they have some novel ability to, say, "unsplice," which humans cannot do. This could potentially allow them to use already made transcripts and then completely change them to produce unique proteins without needing to transcribe another mRNA. This could feasibly allow them to rapidly change what proteins are in each cell (perhaps quick enough that it occurs within the regeneration itself). Although, there would be some instability while now unused proteins get degraded or the splicing/unsplicing ratio stabilizes (the molding period). This would require intense regulation as well as unsplicing and resplicing would now be posttranscriptional, but I digress.
Sorry to bother you with the long post, I just had too many nerdy ideas going through my head. Thanks!
-gallifreyanhotfive
Molecular Biology: 'Unsplicing'
Oh, you thrill me with your biology talk! Molecular biology is not a speciality so apologies in advance for any limited response.
🔬 Lindos and Its Variations
Something to be covered in the new Anatomy and Physiology guide is a wider look at the role of Lindos in Time Lords, so we're hitting the nail on the head here.
Under stress, injury, or during the process of regeneration, the lindal gland significantly increases its production of the hormone Lindoneogen like a caffeine-fueled scientist, resulting in a corresponding surge in lindos cell production. There are several forms of lindos cells, including:
Lindopoetic Progenitor Cells (LPCs): Dormant cells that spring into action upon Lindoneogen stimulation.
Lindopoietic Stem Cells (LSCs): Residing in the yellow bone marrow, ready to differentiate under the guidance of Lindoneogen and the catalytic influence of artron, into ...
Lindoblasts and Phagolindotropes: Specialised cells responsible for regenerating tissue and recycling cellular components from the previous incarnation.
Haemolindocytes: Circulating cells that endow Gallifreyan blood with its regenerative properties.
💡Splicing and Lindoneogen
Lindoneogen could play a key role in alternative splicing, creating specific mRNA isoforms vital for regeneration. This implies that Lindoneogen is not just a cellular signal but also a molecular tool for crafting the necessary protein portfolio for regeneration. So Lindoneogen may trigger the production of specific mRNA isoforms that are vital for the regeneration process, which could lead to the expression of proteins that facilitate the transition of cells into a more pluripotent state.
🖇️Unsplicing
Love this idea. 'Unsplicing' as your concept presents would be particularly relevant during regeneration. It could allow cells to quickly alter their protein expression profiles without the lag of new mRNA transcription. This rapid adaptation would be pretty handy for the efficient transition of cells to suit the requirements of the new incarnation.
🔗Integrating with Lindos Cells
This concept of 'unsplicing' could be particularly prominent in the function of phagolindotropes. As these cells are responsible for consuming the previous incarnation’s cells and replacing them with new ones, their ability to 'unsplice' and rapidly change protein expression would be pretty useful. This mechanism might also support the functions of lindoblasts and haemolindocytes in tissue regeneration and blood adaptability.
🏫 So ...
The addition of splicing and unsplicing mechanisms to the lindos theory suggests a more complex and dynamic process than simple cellular proliferation and differentiation, with dynamic genetic adaptations at the molecular level highlighting the advanced biological capabilities of Gallifreyans. Good work, Batman!
→🫀Gallifreyan Anatomy and Physiology Guide (WIP)
→⚕️Gallifreyan Emergency Medicine/Monitoring Guides
→📝Source list (WIP)
-------------------------------------------------------
》📫Got a question / submission?
》😆Jokes |🫀Biology |🗨️Language |🕰️Throwbacks |🤓Facts
》📚Complete list of Q+A
》📜Masterpost
If you like what GIL does, please consider buying a coffee to help keep our exhausted human conscious for future projects, including complete biology and language guides.
#doctor who#gil#gallifrey institute for learning#dr who#dw eu#gallifrey#gallifreyan biology#gil biology#gallifreyans#ask answered
19 notes
·
View notes
Text

Inspired by the headcannon I sent to @dalekaiken, read DNA Collision if you haven’t yet!
Decided to redo the drawing now that I've gotten a bit more of a handle on drawing digitally. (Click on the image for better image quality.)
The background shows nucleosomes unraveling and I overlaid Hachimoji "ALIEN" DNA code onto Shadow, which represents his nucleosomes rearranging due to chaos energy to reveal his "ALIEN" genes.
8 notes
·
View notes
Text
Histone Modifications
Hello, hello! Today's topic is histone modifications. We are continuing on with the epigenetics theme after my previous educational post about DNA methylation. As described in that post, epigenetics is the study of heritable genetic modifications without a change in DNA sequence (Takuno & Gaut, 2012). Similarly to DNA methylation, histone modifications affect gene expression through regulation of accessibility of the DNA for transcription (Bartova et al, 2008). But before we get into these modifications, let's go over a bit of background information!
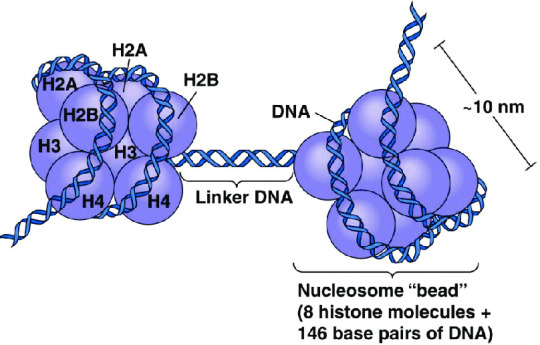
What is a histone, anyway? A histone is a type of protein involved in DNA compaction and organization. In order to fit a genome's worth of DNA into the nucleus of a cell, that stuff needs to be extremely tightly packed! Histones help with this by forming an octomer called a nucleosome, which the DNA wraps around. These nucleosomes then coil together to form a fiber known as chromatin, which goes on to make up a chromosome. When the chromatin is less tightly packed, it is known as euchromatin and it is available for transcription (Bartova et al, 2008). When it is more tightly packed, it is known as heterchromatin, and polymerase proteins cannot access and transcribe the DNA (Bartova et al, 2008). Histone modifications regulate the transition between heterochromatin and euchromatin (Bartova et al, 2008).
(Above image from humanoriginproject.com)
(Above image from Caputi et al, 2017)
The octomer core of a nucleosome is made up of two copies of each of four types of histones: H2A, H2B, H3, and H4 (Marino-Ramirez et al, 2017). Each of these histones includes an N-terminal tail structure, which is the main site of modification (Marino-Ramirez et al, 2017). The tails are modified through addition and removal of certain functional groups or other small structures. Types of modifications include acetylation by histone acetyltransferases, methylation by histone methyltransferases, phosphorylation by kinases, and ubiquitination (Marino-Ramirez et al, 2017). All of this information is used for naming specific histone modifications: Which histone is modified, which amino acid of the histone tail the modification is on, what type of modification is made, and in what amount. For example, H3K9me2 is the name for di-methylation of the 9th Lysine on an H3 histone's tail.
Some important histone modifications and their effects include:
H3K9me2: transcriptional activation + maintenance of CHG DNA methylation in plants
H3K9me3: transcriptional repression
H3K9ac: transcriptional activation
H3K4me1 & H3K4me3: transcriptional activation
H3K27me3: transcriptional repression
H4K16ac: transcriptional activation
H3S10p: DNA replication-related chromatin condensation
(He & Lehming, 2003)
Important Terms: histone, nucleosome, heterochromatin, euchromatin, transcription, epigenetics
4 notes
·
View notes
Text
RNAs, introns and introduction to recombinant DNA.
The genome is a vault of genetic information, but on its own its kind of useless as the information cannot be used in the cell. Utilization of this information requires a coordinated effort from enzymes and proteins, this coordination of chemical reactions is known as genome expression. The initial step of genome expression is known as the transcriptome, a collection of RNA molecules which is made up of the genes active in the cell at the time. (its constantly changing to the environment that the cell is in) the transcriptome is maintained by transcription a simple process in which individual genes are copied and pasted into RNA molecules known as mRNA. The second product within gene expression is known as proteome which you can think of as the cells library of proteins at hand. This gives the cell its unique and individual characteristics that it can express. The proteins in this library known as the proteome are synthesized by the translation of some RNA molecules.
I think it is of utmost importance to start with the most simple and important step…
Transcription:
The first step in transcription is initiation,
Transcription begins with the binding of RNA polymerase 2 to the promoter region of a gene, the most common promotor contains a conserved gene sequence we call the TATA box, but there are many others that exist which are show by the different TF families.
The next step we have the formation of transcription initiation complex, this is when transcription factors such as TATA-binding protein also known as TBP, bind to the promoter, forming a pre-initiation complex. Other general transcription factors join, and RNA polymerase 2 is recruited to form the transcription initiation complex.
Next, we have initiation of transcription where RNA polymerase 2 unwinds the DNA helix at the transcription “start site”. The enzyme catalyses the synthesis of a short RNA primer (which is about 10 nucleotides in length) complementary to the template DNA strand.
The second step is Elongation:
Once the RNA polymerase has synthesized the initial RNA primer, it proceeds to elongate the RNA chain, The RNA polymerase adds ribonucleotide complementary to the template DNA strand. This process is known as RNA chain Elongation.
After this, we have nucleosome remodelling where the DNA in eukaryotic cells is wrapped around histone proteins to form nucleosomes. As transcription proceeds, nucleosomes are temporarily disrupted and then reassembled, allowing RNA polymerase to access the DNA template.
The third step is Termination:
Eukaryotic mRNA molecules undergo post-transcriptional modification, this includes polyadenylation which involves the addition of a poly-A tail to the 3’ end of the RNA. Polyadenylation is then accompanied by cleavage of the RNA precursor at the specific site.
During this process transcription termination signals are recognized by RNA polymerase 2, leading to its dissociation from the DNA template. The termination signal will most likely include the poly-A signal.
The fourth step being RNA processing:
The first step of RNA processing is known as “capping”. Capping the 5’ end of the newly synthesized mRNA is modified with a 7-methylguanosine cap, the whole point of this cap is just to protect the mRNA as it leaves the nucleus and helps in the transportation process.
The second step is splicing, Eukaryotic genes often contain introns (which are kind of gaps or non-coding regions) and exons (which are the important bits, coding regions). In a process called splicing the annoying useless introns are cut and the exons are then pinched together with each other. This is all catalysed by a complex of RNA and proteins known as spliceosomes.
The fifth step being transport and translation:
This is when mRNA is transported out of the nucleus to the cytoplasm where it serves as a template for translation.
The sixth step is regulation:
Gene expression if tightly regulated at the transcriptional level by various elements, including transcriptional factors and chromatin modification, there is also enhancers and silencers that help regulate elements that influence transcription.
We also have post-transcriptional regulation, which is processes such as alternative splicing and RNA stability which play roles in determining the final mRNA products.
It is important to acknowledge that Eukaryotic transcription is more complex than prokaryotic transcription due to the presence of introns, the involvement of multiple RNA polymerases, and the necessity for additional processing steps.
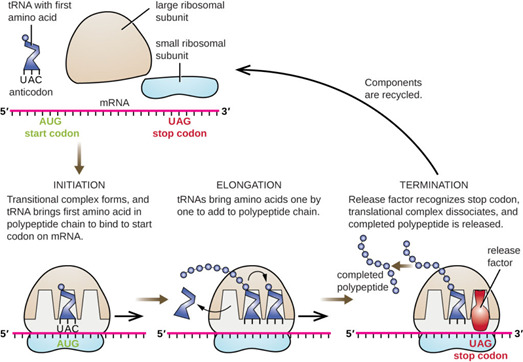
This diagram helps visualise the main steps more simply.
Hope this was clear excuse any mistakes in grammar :)
Referencing:
Picture link above^
2 notes
·
View notes
Text

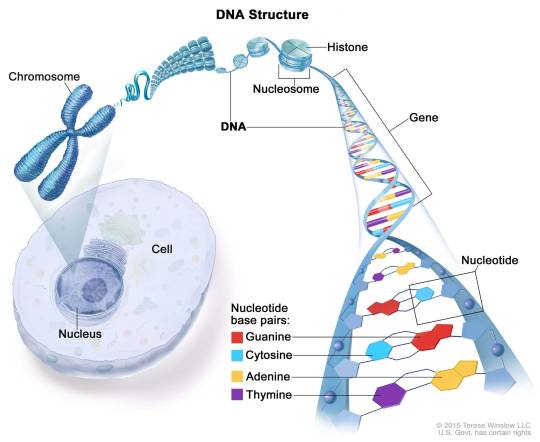
DNA is a molecule that carries the genetic information for all living things. It is made of smaller units called nucleotides, which have three parts: a sugar, a phosphate, and a nitrogen base. There are four types of nitrogen bases: adenine (A), thymine (T), cytosine (C), and guanine (G). The nucleotides are linked together in a chain, forming a strand of DNA. Two strands of DNA pair up with each other, forming a double helix. The pairing is based on the rule that A always binds with T and C always binds with G. This way, the two strands are complementary to each other and can store the same information.
DNA is found inside the cells of all organisms. Most of it is located in the nucleus, where it is organized into structures called chromosomes. Each chromosome contains a long piece of DNA that has many genes. Genes are segments of DNA that code for proteins, which are the building blocks and regulators of life. Some DNA is also found in the mitochondria, which are the energy factories of the cell.
DNA is very long compared to its width. A single strand of human DNA is about 5 feet or 1.5 meters long, but it is only 2 nanometers wide. To fit inside the cell, DNA has to be compacted and coiled. It wraps around proteins called histones, forming beads called nucleosomes. The nucleosomes are further coiled into fibers called chromatin, which can condense into chromosomes during cell division.
If we could stretch out all the DNA in one human cell, it would be about 3 km or 6.6 feet long. If we could stretch out all the DNA in all the cells in our body, it would be about twice the diameter of the solar system. That means that our body contains an enormous amount of information encoded in our DNA. This information determines our traits, such as our eye color and height, as well as our risk of diseases and our response to drugs. It also makes us unique from other individuals, except for identical twins who share the same DNA.
DNA is not static; it can change over time due to mutations, which are errors or changes in the sequence of nucleotides. Some mutations are harmless or beneficial, while others are harmful or cause diseases. Mutations can also create variation among individuals and populations, which is the basis of evolution and natural selection.
DNA is a fascinating molecule that reveals a lot about ourselves and our ancestors. By studying DNA, we can learn more about our health, our history, and our future.
3 notes
·
View notes
Text
Day 19/36

Slept well yesterday. Woke up early today. That set right my mood I have to admit. Completed 2014 AIPMT question paper and also scored well in my daily mock test paper. A mohalle wale uncle passed away today. Please keep them and the family in your prayers tonight🙏

Questions answered: 430
Question of the day:
How many nucleosomes are present in E coli ?
a) Less than 3.3 × 10^6
b) Half of 4.6 × 10^6
c) 0
d) Both a and b




2 notes
·
View notes
Text
nodding when the psychology man says "heritability and nucleosomes" w secret knowledge that shooting range + beer is the only treatment for the aching soul
4 notes
·
View notes
Text
One common type of DNA modification is the methylation of cytosine residues (Figure 2.14A). (...) One, two, or three methyl groups can be added to a single lysine (Figure 2.14B). (...) The resulting gap between nucleosomes is now wide enough for RNA polymerase to bind and initiate transcription (Figure 2.14C). (...) The chromatin structure is then "remodeled" in an ATP-requiring reaction and subsequently methylated, resulting in tighter condensation and heterochromatization of the DNA region involved (see Figure 2.14). (...) Polychrome group protein complexes include multiple forms of Polycomb repressive complex 2 (PRC2), which catalyzes methylation of histones, which are components of nucleosomes whose methylation tends to inhibit transcription of the associated DNA (see Figure 2.14).
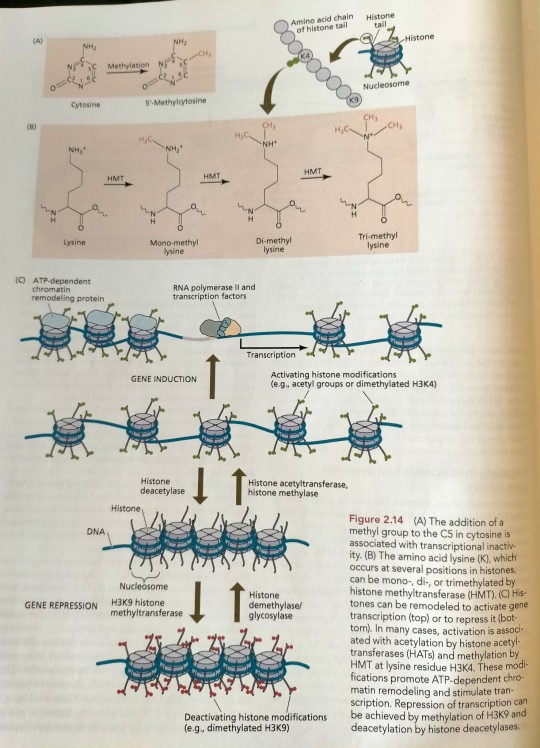
"Plant Physiology and Development" int'l 6e - Taiz, L., Zeiger, E., Møller, I.M., Murphy, A.
#book quotes#plant physiology and development#nonfiction#textbook#chromatin#chromosomes#genes#genetics#transcription#dna#rna#genetic modification#Polycomb repressive complex#prc2#polychrome#methylation#nucleosomes
0 notes
Text
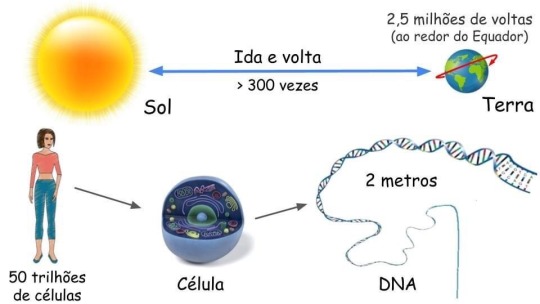
O TAMANHO DO NOSSO DNA
O DNA (ácido desoxirribonucleico) é a molécula que armazena a informação genética para o desenvolvimento e funcionamento de um organismo. Ele é feito de duas fitas ligadas que se enrolam uma na outra, assemelhando-se a uma escada torcida – uma forma conhecida como dupla hélice.
Cada célula humana contém cerca de 2 metros de DNA. Estima-se que o corpo humano tenha cerca de 50 trilhões de células - o que corresponde a 100 trilhões de metros de DNA por humano.
Considerando que o Sol está a 150 bilhões de metros da Terra. Isso significa que cada um de nós tem DNA suficiente para ir daqui até o Sol e voltar mais de 300 vezes ou dar 2,5 milhões de voltas em torno do Equador da Terra!
REFERÊNCIA
https://www.nature.com/scitable/topicpage/dna-packaging-nucleosomes-and-chromatin-310/?fbclid=IwAR1zPdV8_VaWKfst-oVZNICPal3Q41rLamStNemKtSbviMVxh0iPTW2OuaM
1 note
·
View note
Link
0 notes
Text

Inspired by the headcannon I sent to @dalekaiken, read DNA Collision if you haven't yet!
The background shows nucleosomes unraveling and I overlaid the Hachimoji ALIEN DNA code onto Shadow.
And to prove that I'm definitely more comfortable with traditional media than digital, I've included my practice sketches (I had a full digital version before this one I hated and then redid this one like 3 times and I'm still not at all comfortable with digital yet).


#shadow fanart#shadow the hedgehog#still not happy with it#but I don't have the time or energy to try again lol
8 notes
·
View notes
Text
Refined mechanism of promoter Nucleosome-Depleted Regions resetting after replication
BioRxiv: http://dlvr.it/T5W8t4
0 notes