#protein sequence
Note
NGN!!
Next generation network?
Or amino acids
N-G-N
Asparagine-Glycine-Asparagine
I have found a protein with several NGN repeats in Haliotis asinina, is that what you meant? Does that mean you are indirectly calling me an 'ass'?
#is this some code#roleplay#rp#very short amino acid sequence#but if this is true a well thought out insult#sherlock roleplay#sherlock rp#protein sequence#ngn#that was the first protein result while googling NGN
6 notes
·
View notes
Text
the only thing my sister and i have to offer to the rise fandom is theories about how lou jitsu's dna recombined with the turtles
#i think about this a lot. like an embarrassing amount actually#i also think about what sorts of enzymes draxum mustve thrown into the ooze to trigger it all#i think there was probably a mystic factor as well#like a mystic catalyst#the turtles (and now splinter) would be considered transgenic organisms#and i dont have a whole lot of background in transgenic techniques so its a little harder for me#there are a lot of ppl at work who are experts on making humanized mouse models so i should ask them LOL#most of my research is on rna and protein expression and not dna modification#still compels me tho#megan we should try to find if anyone has sequence aligned turtle and human dna#im curious how similar the genomes are#and also what genes and consensus sequences are shared between reptiles and humans
6 notes
·
View notes
Text
my cell bio professor just referred to an example protein signal sequence as “non-canonical” and I’m kind of obsessed with it. yeah I really hate the canon Akt pathway but omg the fanon version is so good
6 notes
·
View notes
Text
Ten Hacks for Predicting Protein Properties from Sequences using Machine Learning
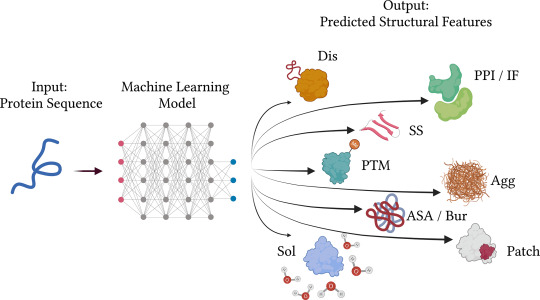
The success of machine learning-based approaches for the prediction of protein properties from their amino acid sequences can be attributed to the overall accessibility of genome sequencing data. One of the most crucial challenges for bioinformaticians is still predicting the functional properties of proteins. Ten issues or guidelines that reflect best practices, especially for techniques that predict the functional, structural features of proteins using protein sequence data as input, are shown below to help developers overcome this gap:
Sequence-based approaches often perform worse than those based on structures. There has been significant development in the field of structure prediction during the past several years, which has coincided with the expansion of empirically determined structures. Particularly, AlphaFold2 represents a significant advancement in structure prediction. Many significant protein types and areas still lack access to accurate structural data, nevertheless. Furthermore, it hasn’t been shown yet and could still be fairly restricted how valuable predicted structures are as input for the prediction of functional features like interface areas or binding sites. Structure-based functional property prediction is not always more precise than sequence-based methods. In order to close this gap, the area of sequence-based prediction of protein function and structure features was created.
Continue Reading
46 notes
·
View notes
Text
what if I reverse engineer a post that can get protein blasted into one of my peptides. yes it's cheating but consider. it would be so cool if I could pull it off
#the sequence doesnt start with a methionine tho (cause its a peptide) so idk if the protein blast blog would catch it.....#but my full protein is 306 amino acids long... thats over 900 nucleotides.... no way in hell am i making a post that long sldjdk#edit: my dumbass forgot my protein is cleaved from a larger orf anyways. it starts with a serine.#truly i will never be recognized by protein wizard 😔
2 notes
·
View notes
Text
Central Dogma of Life
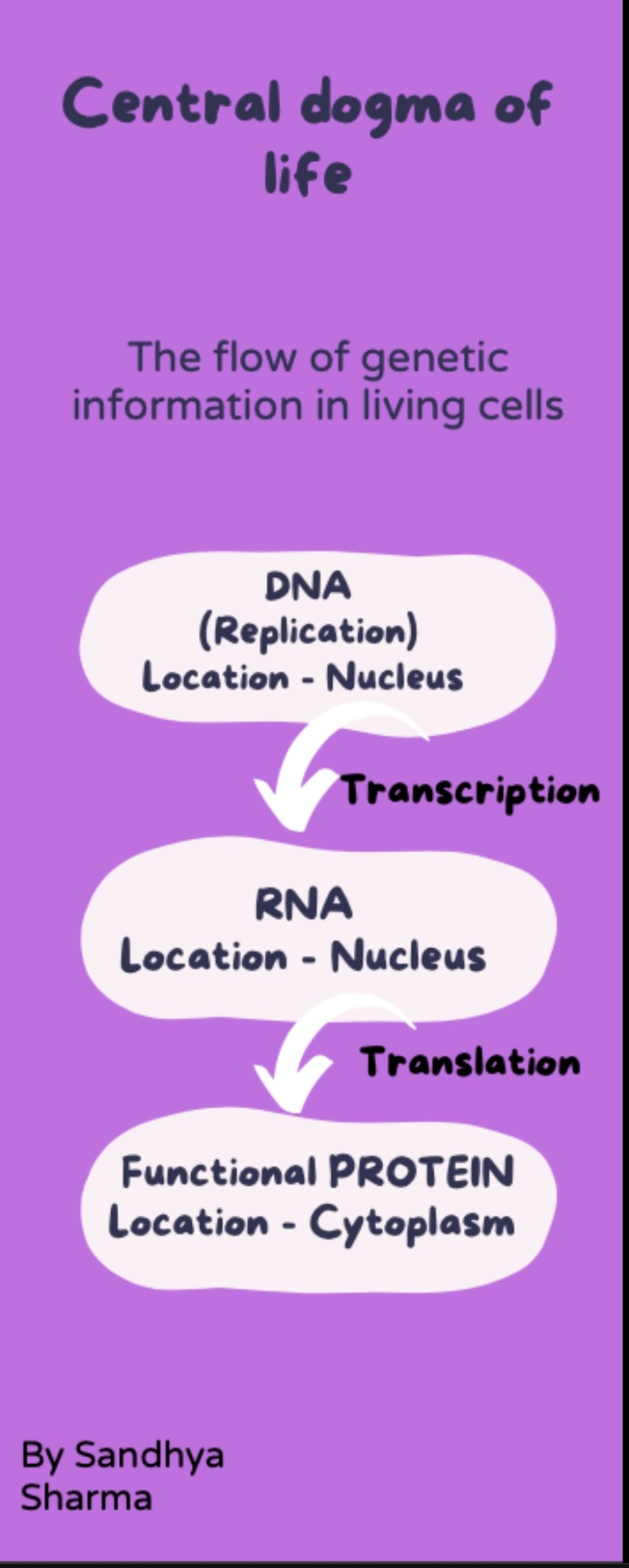
#dna#chemistry#science#dna activation#biotechnology#biochemistry#biology#rna#rna sequencing#replication#protein#infographic#useful#resources#info post#information#helpful#science communication#science class#life science#genetics#cells at work
1 note
·
View note
Text
https://www.merexpression.com/read-blog/141307_protein-sequencing-market-size-share-and-forecast-2031.html
The Protein Sequencing Market in 2023 is US$ 1.4 billion, and is expected to reach US$ 1.87 billion by 2031 at a CAGR of 3.70%. FutureWise Research published a report that analyzes Protein Sequencing Market trends to predict the market's growth. The report begins with a description of the business environment and explains the commercial summary of the chain structure. Based on the market trends and driving factors presented in the report, clients will be able to plan the roadmap for their products and services taking into account various socio-economic factors.
0 notes
Text
Protein Sequencing Market Size, Volume, Demand, Outlook and Forecast Year (2024-2033) | BIS Research

In the ever-evolving landscape of life sciences and biotechnology, the Protein Sequencing Market stands at the forefront of groundbreaking advancements. As scientists continue to delve into the intricacies of proteins, the demand for accurate and efficient protein sequencing methods has surged. This blog aims to shed light on the current trends, challenges, and future prospects of the Protein Sequencing Market.
Proteins, the building blocks of life, play crucial roles in various biological processes. To comprehend their functions and unlock their potential in fields like medicine and agriculture, scientists turn to protein sequencing. This process involves determining the precise order of amino acids in a protein chain.
Empower Your Strategies: Receive Your Sample Report and Conquer the Protein Sequencing Market @ https://bisresearch.com/requestsample?id=1724&type=toc
Key Market Drivers:
Biopharmaceutical Research: With the rise of biopharmaceuticals, there's an increased need for in-depth protein analysis. Protein sequencing is instrumental in drug development, ensuring the efficacy and safety of therapeutic proteins.
Disease Diagnosis and Treatment: Protein sequencing is pivotal in understanding diseases at the molecular level. Accurate identification of proteins associated with various ailments facilitates the development of targeted therapies and personalized medicine.
Technological Advancements: Continuous innovations in sequencing technologies, such as mass spectrometry and next-generation sequencing, have enhanced the speed and accuracy of protein sequencing, driving market growth.
Challenges in the Protein Sequencing Market:
Cost Constraints: High initial costs associated with advanced sequencing technologies can be a barrier for smaller research institutions and laboratories.
Data Analysis Complexity: The vast amount of data generated during protein sequencing requires sophisticated bioinformatics tools for analysis, posing a challenge for researchers unfamiliar with these technologies.
Sample Complexity: Some proteins are inherently challenging to sequence due to their size, post-translational modifications, and structural intricacies, creating hurdles for researchers.
Single-Cell Protein Sequencing: The ability to sequence proteins at the single-cell level opens new avenues for understanding cellular heterogeneity and disease mechanisms.
Integration of Artificial Intelligence: AI is increasingly being incorporated into protein sequencing workflows for data analysis, pattern recognition, and prediction, expediting the research process.
Advancements in Mass Spectrometry: Ongoing improvements in mass spectrometry technologies contribute to faster and more accurate protein sequencing, revolutionizing the field.
Future Outlook:
As the Protein Sequencing Market continues to expand, collaborations between academia
0 notes
Text
How to Use Protein Data Bank (PDB)? A Complete Guide!
Protein Data Bank Homepage
In this comprehensive guide, we’ve explored navigating the Protein Data Bank (PDB) and efficient protein structure searches. We’ve covered various search methods like keywords, PDB IDs, advanced and sequence searches, and structure similarity searches. We’ve also highlighted using PDB IDs, advanced features, and multiple access channels like the PDB website, FTP, APIs,…
View On WordPress
#how access pdb#how to search protein data bank#how to use pdb#methods for protein search#pdb advanced search#pdb ID search#pdb keyword search#pdb result page navigation#pdb sequence search#pdb structure similarity search#protien structure#where to find protiens
0 notes
Text
No single DNA sequence or binding protein is common to all phytochrome-regulated genes.
"Plant Physiology and Development" int'l 6e - Taiz, L., Zeiger, E., Møller, I.M., Murphy, A.
#book quote#plant physiology and development#nonfiction#textbook#dna sequence#binding protein#phytochrome#gene regulation
0 notes
Text
MicroRNA
Working of miRNA
MicroRNA (miRNA) consist of a family of molecules that helps cells to control the kinds and amount of proteins. In other words cells use these molecules to help in controlling gene expression of a particular gene in making too much or too little or the normal amount of protein at a particular time. Protein expression helping to know how cells use gene’s DNA to make protein.…

View On WordPress
#2023#DNA#DNA Sequencing#Dr. Suresh Kaushik#gene expression#gene silencing#GingerFingers#MicroRNA#miRNA#mode of action#molecules#NGS#protein synthesis#RNA
0 notes
Link
Protein Sequencing Market Share Past, Present Data and Deep Analysis 2022-2030
0 notes
Text
Macvector protein sequence

Overall sequence similarity with the mouse protein is shown on the right. The percent similarity of the SAM domains and other regions to the corresponding regions of the mouse protein is shown. (D) Schematic comparison of the amino acid sequences for mouse, rat, human, chick and zebrafish mr-s proteins. Branch lengths reflect the mean number of substitutions per site. Amino acid sequences were analyzed by the neighbor-joining method in MacVector 7.2. (C) Phylogenetic tree of SAM domain-containing proteins. The sites that were targeted for mutagenesis are indicated by arrows. Conserved amino acid residues are shown with a dark shadow and functionally similar residues are shown with a light shadow. (B) Alignment of SAM domain sequences for SAM domain-containing proteins. The underline indicates a putative polyadenylation termination signal. Boxed amino acids are the SAM domain sequence and the dashed box indicates a putative nuclear localization signal. (A) mr-s nucleotide and amino acids sequences. In addition, the finding of rare heterozygous QRX sequence changes in three individuals with retinal degeneration raises the possibility that QRX may be involved in disease pathogenesis.Mr-s nucleotide and amino acid sequences. These results indicate that Qrx may be involved in modulating photoreceptor gene expression. Nonetheless, a 5.8 kb upstream region of human QRX is capable of directing expression in presumptive photoreceptor precursor cells in transgenic mice. Qrx is present in the bovine and human genomes, but appears to be absent from the mouse genome. As predicted from the amino acid sequence homology, recombinant DiTG catalyzed. QRX synergistically increases the transactivating function of the photoreceptor transcription factors Crx and NRL and it physically interacts with CRX. protein ERp60, a PDI isoform found in the lumen of endo- plasmic reticulum. Although Qrx and Rx/Rax show similar DNA binding properties in vitro, the two proteins demonstrate distinct target selectivity and functional behavior in promoter activity assays. Its homeodomain is nearly identical to that of Rx/Rax, a transcription factor that is essential for eye development, but it shares only limited homology elsewhere. Qrx is preferentially expressed in both the outer and inner nuclear layers of the retina. Ī novel paired-like homeobox gene, designated as Qrx, was identified by a yeast one-hybrid screen using the bovine Rhodopsin promoter Ret-1 DNA regulatory element as bait. Six microliters (20 ng/ml) purified Scc1 or Scc1-P in H100 GIBBS sampling option of the MACAW program (Schuler et al., 1991 was added to. Multiple sequenceScc1 cleavage-competent yeast extracts after overexpression of alignment construction and analysis, with statistical evaluation ofEsp1 and control extracts were obtained as described (Uhlmann et the significance of motif conservation, were performed using theal., 1999). to the MacVector 6.5. Database screening for amino acid patterns was performed using the PATTINPROT pro-Cleavage of Purified Scc1 in Yeast Extracts gram at the NPS2 server (Combet et al., 2000). The L-Zip motif sequence and the confirmation of the two helices were found by submitting the nad4L amino acid sequences we obtained for several schistosome spp. the PHI-BLAST program (Zhang et al., 1998). Additional database searches combining BLASTP with a pattern analysis were performed usingmetaphase-like phosphorylated Scc1. This protocol yielded 25 mg of purified Scc1 or purified BLAST program (Altschul et al., 1997). The final eluate was dialyzed against buffer H100 and concentrated bytion by cleaving other proteins or by a different mechaCell 384 ultrafiltration. Whether Esp1 performs this func- as Hsp70 from the insect host cells by mass spectrometry. 96a Ciosk et al., 1998 contaminating protein that copurified with Scc1 and was identified Kumada et al., 1998). To access GenBank and its related retrieval and analysis services, go to the NCBI home page at: Complete bimonthly releases and daily updates of the GenBank database are available by FTP. BLAST provides sequence similarity searches of GenBank and other sequence databases. GenBank is accessible through NCBI's retrieval system, Entrez, which integrates data from the major DNA and protein sequence databases along with taxonomy, genome mapping, protein structure and domain information, and the biomedical journal literature via PubMed. Daily data exchange with the EMBL Data Library in the UK and the DNA Data Bank of Japan helps ensure worldwide coverage. Most submissions are made using the BankIt (web) or Sequin program and accession numbers are assigned by GenBank staff upon receipt. GenBank (R) is a comprehensive database that contains publicly available DNA sequences for more than 140 000 named organisms, obtained primarily through submissions from individual laboratories and batch submissions from large-scale sequencing projects.

0 notes
Text
De novo protein sequence analysis

Oxford Nanopore and Illumina MiSeq libraries were prepared using a rapid sequencing kit (SQK-RAD004, Oxford Nanopore Technologies: Oxford, UK) and TruSeq Nano DNA kit (Illumina, San Diego, CA, USA), in accordance with the manufacturers’ instructions. We also identified mechanical stimulus-responsive genes and their functional annotations which implied melanin biosynthesis genes are up-regulated to introduce browning phenotype and fatty-acid ligase genes to be up-regulated in A. In summary, through the combination of de novo genome assembly and transcriptome analysis, we assembled the first pseudo-chromosome-level genome of East Asian A.bisporus. Meanwhile, we also identified up-regulated melanin biosynthesis pathway genes at an early time point, but they were less enriched later and were expected to produce DOPA-melanin or catechol-melanin. In particular, fatty-acid ligase genes were significantly up-regulated in both the GO analysis and the gene set enrichment analysis (GSEA) throughout all analyzed time points. We identified many differentially expressed genes (DEGs), and among those, many gene ontology (GO) terms were enriched throughout the mechanical stimulus. bisporus, we applied a simple mechanical stimulus and performed transcriptomic analysis in a time-course manner. To identify transcriptomic impacts of mechanical stimulus in A. Thus, we confirmed that strain KMCC00540 could be used as a representative cultivar in a study of A. bisporus strain KMCC00540 shares most genomic blocks and similar genomic structures with the European strains H97 and H39. Through comparative genomics analysis, we could identify that A. bisporus through the de novo assembly of the strain KMCC00540. We identified the genome of our cultivar as a standard genome of A. bisporus and identified which genes respond to a mechanical stimulus, which provided key hints for improving the post-harvest biological control of A. In summary, we assembled the chromosome-level genomic information on a Korean strain of A. Mechanical stimulus induces up-regulation in long fatty acid ligase activity-related genes, as well as melanin biosynthesis genes, especially at early time points. To identify which genes respond to a mechanical stimulus that induces browning, we performed a time-course transcriptome analysis based on the de novo assembled genome. bisporus suffers from browning even from only a slight mechanical stimulus during transportation, which significantly lowers its commercial value. A comparative genomic analysis with cultivar H97 indicated that most genomic regions and annotated proteins were shared (over 90%), ensuring that our cultivar could be used as a representative genome. The KMCC00540 genome had 13 pseudochromosomes comprising 33,030,236 bp mostly covering both strains. After generating a scaffold-level genomic sequence, we inferred chromosome-level assembly by genomic synteny analysis with the representative A. We performed de novo genome assembly with a South Korean white-colored cultivar of A. Agaricus bisporus is one of the world’s most popular edible mushrooms, including in South Korea.

0 notes
Text
https://www.cienciared.es/blogs/210261/Protein-Sequencing-Market-Size-Share-and-Forecast-2031
The Protein Sequencing Market in 2023 is US$ 1.4 billion, and is expected to reach US$ 1.87 billion by 2031 at a CAGR of 3.70%. FutureWise Research published a report that analyzes Protein Sequencing Market trends to predict the market's growth. The report begins with a description of the business environment and explains the commercial summary of the chain structure. Based on the market trends and driving factors presented in the report, clients will be able to plan the roadmap for their products and services taking into account various socio-economic factors.
0 notes