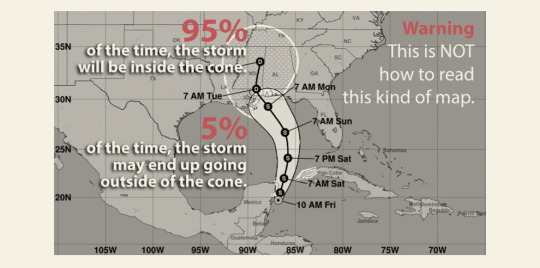
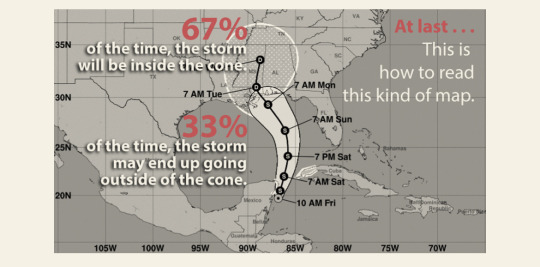
Text
Neural Networks
feedforward, fully-connected neural networks
composition of a neural network
input node (s) : put the data into the neural network
hidden node:
linearizes the data with (weight • x + bias) = z
warps that linearized data with a nonlinear factor known as sigma with (z • sigma)
returns an activation output a that is then passed to the next hidden node as its input
hidden node weights are written atop the lines connecting one node to another
the last node is the output node whose activation is churned out as the y-hat prediction
because each input is the output of the last node, we could theoretically plug them all in by hand
general flow of the neural network
forward propagation – feeding activations into the next node »»»»
loss function – the error that is generated with the output of the forward propagation
back propagation – the minimization of the loss function via altering the nodal weights/biases
where do networks get deep and wide?
depth is layered growth and width is growth along the same layer
depth = parent + many child nodes and width = many sibling nodes
a fully-connected neural network is a network in which every node is connected to all of the nodes in the layers above and below
(but not between sibling nodes because info flows layer-by-layer)
express all of these weights associated with the connections in matrices and linear algebra in order to visualize it efficiently
training assigns useful or non-useful weights to each of the connections in a fully-connected neural network
the process of using a neural network
input node(s) can take on many different dimensions of data to run through the hidden nodes. the general form of your input shape is the number of dimensions/traits you have in your X-matrix
forward propagation training: first start with randomized weights and run the data through the neural network
back propagation training: for a single layer, take the derivative of the loss function with respect to each of the nodal weights (gradient descent!) in order to minimize loss and find its sensitivity to the given random weights. So with 12 nodal weights, the gradient descent would be calculated across a 12-element vector
back propagation training: all leftover error from a single layer back propagates to the layer preceding it and then new estimates are generated forward, so that error back propagates and estimates forward propagate
the determination of the output node's dimensions is super critical to the proper interpretation of the NN
the output shape of an NN is often composed of a probability distribution of all possible labels (i.e., the % likelihood that the datapoint belongs to each label)
often a little output layer is created to generate (6)
the loss function is a measure of error – but actually there are many different loss functions that can be utilized
the optimizer is that which is utilized to minimize the given loss function (i.e., gradient descent)
for large networks and neural networks
normal gradient descent is too computationally heavy to run on all of these nodes and datapoints
so, the dataset is split into groups called batches of data for ease of updating weights iteratively, and an epoch is one round of training with the entire dataset
and stochastic (random) gradient descent is utilized to train the neural network instead
you can also add different algorithms to your SGD to further improve its efficacy (there are many)
3 notes
·
View notes
Text
Short-notes: K-Means Clustering
unsupervised learning method
iteratively update random clusters based on distances from the datapoints in each cluster
E-step: "how many datapoints are closest to this centroid and not to the other centroids?" assign each random centroid to a cluster
M-step: "how do I reduce the distance between the cluster and its newly assigned centroid?"
6 notes
·
View notes
Text
Genome Wide Association Studies (GWAS)
why genome-wide studies?
looking at the genotype can reveal useful information about a disease
i.e., use genotypes to develop treatments related to gene expression inhibition or upregulation
other uses: personalized/custom treatments, disease risk predictions for screening & recommendations for lifestyle changes
types of genomic associations
information varies based on nature of genome-disease relationship
mendelian disease: complete penetrance w/ categorical + or - disease phenotype, single mutational cause, clear inheritance patterns
complex trait disease: quantitative + or - disease phenotype influenced by multiple gene loci & environment in a possibly non-additive manner
linking loci to complex traits/diseases
categorical linking via association mapping: split a sample group into the case and control groups and look at allelic variations between the two
find the significance of your allelic differences via 1) odds ratio and 2) chi-squared analysis
chi-squared = ∑ (observed-expected)^2 / expected, where expected = total cases in case group • control group's percentage of a certain allele
quantitative linking via regression analysis: (y) phenotypic trait v. (x) allelic variation
collection of box & whisker plots for each allelic variant, that is then placed into a linear regression trendline
GWAS linking via association mapping across SNPs
use a Manhattan plot (-log p-value v. SNP's chromosomal position) to visualize associative relationship between disease p-value & SNP
utilize correction methods (Bonferroni & Benjamini-Hochberg) to lower false positives with thousands of SNPs across the genome
can also utilize microarray analysis with specifically significant SNP loci
Not all SNPs across the genome need to be looked at, however
SNPs may be in non-coding regions, or SNP linkage w/ other SNPs/the disease phenotype may not be perfect
SNP linkage falls across a sliding spectrum of equilibrium that affects the association between a SNP and the observed phenotype
challenges to GWAS
distinguishing between causal & linkage disequilibrium linkage between SNPs and disease phenotype
finding and observing rare SNP linkage to diseases
heritable variation (h^2) at quantitative trait loci (QTL, or loci that do contribute to a complex trait) is around 1-50%
new innovations for the future
use machine learning to consider different variables as you determine which SNPs are causal
i.e., determine likelihood that the SNP at hand is causally linked to a disease based on the environment it rests in
0 notes
Text
Principle Component Analysis
unsupervised method that reduces the dimensional space of a dataset (gives it some structure)
PCA creates a space for/version of the X-dataset with redundant correlations minimized, that can then be utilized by other ML algorithms
while linear regression's error is minimized with respect to the x-axis, PCA's error is minimized with respect to the trendline itself (think diagonal)
PCA: find the slope of the trendline that minimizes the error relative to the line itself
projection: drop all of the points onto whatever axis or trendline. minimal variance of projected points = large error of actual datapoints' y-values
PCA = find maximum variance of PROJECTED POINTS and thus minimum variance of ACTUAL y-VALUES. the line that gives this is the first principle component
the centroid is that which the projection variance/trendline is rotated around to find the line of maximum projected variance
to sum:
PCA can be defined in one of two ways:
find the trendline that gives the least dataset error relative to said line (rather than the x-axis)
find the trendline that gives the most dataset variance when the dataset is projected onto said line
» and finding the maximum projected variance also finds the minimum dataset error relative to the trendline
» the trendline is known as the first principle component and the mean squared error of that FPC is the explained variance
after the FPC – second principle component
each additional principle component must be perpendicular (orthogonal if in multiple dimensions) to the others
find the orthogonal SPC that gives you the best analysis of the dataset
the SPC takes the variance that falls outside of the FPC's explained variance and explains it, wrapping up all remaining variance
the distances between a given point and the FPC/SPC are known as the eigenvalues of each, and capture the relative amounts of variance that each component is able to explain
PCA allows for as many principle components as there are dimensions in/aspects of your X-dataset matrix
on the whole, the dataset may have been collected in terms of feature A and feature B, but may be best described as a collection of linear combinations AB and BA (FPC, and SPC)
graphing PCA
take the eigenvalue vector directions and make them the new axes. plot the datapoints along those component eigenvalue vectors.
the farther your points are along this new graph, the better – the distance is a measure of the efficacy of your PCA compression
so in this way, PCA is an unsupervised method that acts as a dimensionality reducer
so, why reduce the dimensionality of the dataset?
because the more correlated your individual X-matrix features are, the less useful they are (i.e, 2 or more features tell you the same thing about the y-outcome)
so PCA reduces those dimensions/features that give us redundant correlations by mapping the dataset only along the principle components
still, no information is lost because the full dimensionalities of the dataset are required to form the PCA in the first place.
» this is known as the manifold hypothesis! i.e., the dataset's relationships exist on a lower dimensional subspace than that which it was collected in
explained variance efficacy
a dataset has a total variance, some of which PCA attempts to explain in reducing the X-matrix's dimensionality
the space in which the dataset was collected generates certain relationships that have certain variances
by optimally altering this space and reducing dimensions the PCA is able to change those variances – explaining them through new relationship depictions
the more variance the PCA is able to explain, the better!
[cumulative explained variance plot]
the cumulative explained variance v. number of components relationship gives the efficacy with which a relevant PCA can capture variance
this is known as a cumulative explained variance plot and returns how many original X-dataset matrix dimensions can be discarded without losing explained variance
discarding dimensions and reducing the space of the X-dataset via PCAdecreases the likelihood of your algorithm overfitting to a complex/detailed dataset
7 notes
·
View notes
Text
Ensembles
fancy word for averaging (good for reducing variance!)
majority voting classifier
multiple models make separate decisions, which are then counted and the majority decision is the final output
hard voting: singular decision, with each model only returning a yes/no to the final output
in hard voting, the model is NOT allowed to give a certainty probability value of its decision, a la logistic regression classification
soft voting: each model gives a vector array [P(class 0) P(class 1) etc etc] and the final output averages the models' probabilities for each classification category before the "vote"
in soft voting, you can place weights on each of the models so that if a model has been right a lot of the time, it gets more of a say in the final vote » this is the idea behind boosting
bagging
bootstrap [make the bags] aggregating [train the models on the bags], or creating different datasets from your original in order to generate ensembles
create a bag from your original data by sampling from your dataset with replacement (aka bootstrapping); several bits of data are always left out of the bag
make a lot of bags and fit different models to them, and these become your ensemble of predictions that you take the final vote from!
a random forest is this process, with the models used being decision trees!
bagging decision trees takes the low-bias, high-variance decision tree model (a deep tree) and DECREASES the high variance! i.e., addresses the decision tree problem set
boosting
takes stumps (high-bias, low-variance small power decision trees) and builds them into trees sequentially
conglomerate the stumps into a tree, boosting the total effect by training each stump in sequence
steps to boosting:
all datapoints carry a uniform weight at the beginning
a stump classifies the datapoints. decisions that are correct get an increased weight
the next stump in the sequence now gets the datapoints with their new weights. it classifies the datapoints.
loop classifications using all the stumps in the sequence, with each next stump receiving the weighted datapoint classifications from the last loop's stump
all the models share the same updated weights, which is essentially how this "tree of trees" communicates.
random forest
two components:
bagged data with each bag being fed into a different tree
from one bag, only a SUBSET of the X-matrix is shown to a single node
(i.e., if X has 10 features, two random columns of features are shown to node A and two different random columns are shown to node B)
[then of course you aggregate and majority hard/soft vote at the end. scikit-learn uses soft voting and then an ensemle-level thresholding at the end]
why do this? to further prevent overfitting! i.e., if you underfit a single node, you can balance the overfitting of the entire tree and reduce the variance
forest evaluation – without a test set, use out of bag error i.e., test the data with unincluded dataset portions from each bag
0 notes
Text
Evolutionary Comparative Genomics
unified evolutionary theory
modern synthesis of historic ideas about evolution
Darwin: natural selection
Mendel: mendelian inheritance
population genetics: neutral evolution, genetic drift
tenets of unified evolutionary theory
undirected mutation and recombination gives rise to population variation, which is then pushed here and there by evolutionary pressure
∆gene frequency results from: genetic drift, gene flow (gene transfer between populations of the species), & natural selection itself
most adaptive mutations generate slight phenotypic changes, and actual alteration occurs slowly through accumulated mutations
cladogenesis: diversification arises through speciation due to gradual reproductive isolation between populations of a single species
higher order taxonomy arises with sufficient time to evolve
evolutionary genetics: how genomes differ between species (macroevolution) and within species (microevolution)
inferring gene age
a phylogenetic tree between different species can be utilized to compare the ages of two genes
the gene age can then be approximated via either the fossil record or the mutational rate
i.e., MYH16 gives most primates strong jaw muscles but humans have an inactivating mutation in MYH16 – so we can approx its age
gene function and evolutionary rate
catalytic gene function preceded regulatory gene function, historically
for cancer: caretaker genes maintain DNA integrity, gatekeeper genes regulate the cell cycle/proliferation (your oncogenes and tumor suppressors)
if two species share a gene and one species shows an accelerated rate of evolution in that gene, the gene's responsibility for that species' phenotypic differences may be indicated
use dN/dS for expressed genes because it involves codons; there are other measures of mutation rates in intronic regions
0 notes
Text
Decision Trees
Decision tree: a mechanism for making decisions based on a series of "if" factors
Root node – the decision that matters the most, ergo the decision that precedes all others
Leaf node – the last outcome of a decision tree branch
format: if (node1) and (node2) and (node3)...and (node n), then (leaf node: final decision)
determining the root node: intuitive in a simple human context BUT with data, one must use an algorithm
utilize mechanisms from previous studies (i.e., data visualization with histograms, regression) to find one that gives the most classifying information
each node is "greedily constructed," i.e., the model extracts as much information as it can for an individual node without thinking about info that could be derived from future nodes
so the decision tree algorithm is run recursively, again and again, until the data is sifted perfectly into individual categories
you can use the same features over and over again (recursive!!) to separate out stat at different levels of "purity"
each time there is new data, you can run them through the tree and observe where they end up!
growing a tree
split the parent node and pick the feature that results in the largest "goodness of fit"
repeat until the child nodes are pure, i.e., until the "goodness of fit" measure <= 0
preventing tree overfitting
overfitting: too pure, too in-depth tree, too many nodes that are specifically fitted to the given dataset rather than the real-world function
set a depth cut-off (max tree depth)
set a min. number of data points in each node
stop growing tree if further splits are not statistically significant
and cost-complexity pruning: utilizing some alpha regularization threshold to prevent overfit
cost-complexity pruning
regularizing the decision tree via alpha at the cost of tree purity
so as alpha increases, impurity also increases. number of nodes and depth of tree decrease
real-world accuracy increases to a certain point before decreasing – as prevention of overfitting with regularization becomes underfitting
1 note
·
View note
Text
Detecting Evolutionary Selection in the Genome
natural selection: an organism that is better adapted to the environment will survive and reproduce
natural selection of a trait requires the following:
heritable variation of the trait
differential reproduction of the trait
types of selection:
negative selection: stabilizing/purifying selection AGAINST variation from some norm or conserved/constant original
positive selection: directional selection FOR variation from the norm (see: environmental change as a selection factor)
neutral evolution: trait becomes dominant/common via random chance, i.e., no selective pressure pushes possession of this trait. allows mutations to freely occur
Kimura's neutral theory of evolution: most evolution is neutral!
in the noncoding region of the genome, this especially holds because these areas are not expressed phenotypically!
in the coding region of the genome, neutral and deleterious mutations will be more common (enhancing/beneficial mutations are RARE)
detecting selection in the genome
why detect? map evolutionary history, predict disease selection, and identify functional regions of the genome
how to detect selections? use the d(n)/d(s) ratio
d(n) = number of observed mutations ÷ number of possible mutations for non-synonymous mutations that alter the corresponding amino acid
d(s) = the above but for synonymous mutations that do NOT alter the corresp. amino acid
for neutral evolution, d(n) = d(s) and omega=1
in positive/directional selection, omega > 1 | in negative/stabilizing selection, omega < 1
need to account for POSSIBLE as well as OBSERVED single-point mutations, n.s. and s., to correct for different mutational possibilities
assumptions made in the initial modeling for selection in the genome
lack of accounting for codon bias in some organisms, which leads to an overestimate of s mutations
sampling bias towards individuals and sample groups analyzed
synonymous and non-synonymous mutations are not equally likely! different mutation locations and pathways will produce various ns/s ratios
transitions are more likely than transversions (T-C, G-A) – solve resulting underestimation of s mutations via k = transitions:transversions = 2 for whole genome and 3 for coding regions
maximum likelihood estimation
i.e., which detected selection model is correct??
MLE allows comparison of various evolutionary models/parameters
use MLE to find the evolutionary selection model that has the highest likelihood of generating the observed mutations
you will need!
observations to be mapped by a model
evolutionary model to be tested
a method for computing the likelihood of getting mapped observations with tested evolutionary model
the math of things involve the following symbols:
k = transition:transversion ratio
π(j) = codon usage for any given codon j
omega = d(n)/d(s) from above
q(i, j) = the rate of transition from codon i to codon j
calculate the rates of a transition between different codons w/ a single point mutation:
q(i, j) = 0 for > 1 mutation
= π(j) for syn. transversions
= π(j) • k for syn. transitions
= π(j) • omega for nsyn. transversions
= π(j) • k • omega for syn. transitions
site or branch-specific selection detection
conduct an analysis on whole genome/gene + whole tree with the assumption that selection is equal across gene/tree
» this assumption of equivalent selection across the genome is often mistaken, though :(
most mutations are either neutral or deleterious, so generally across an averaged genomic selection analysis the omega is = 1 or < 1
» this purification favoring trends towards missing the rare positive solutions when conducting a genome-wide analysis of selection rates
looking at the whole tree generally requires long time scales; there are other models, however, that can vary the time scale of the selection analysis
» McDonald-kreitman test (MKT) – calculate an omega within species [substitutions] via short time scale; calculate omega between species [SNPs] via longer time scale
0 notes
Text
Confusion Matrix!
a box that contains the 2 correctly and 2 incorrectly classified values from a model | i.e., [true, false] x (predicted-true, predicted false)
<^ and v> are true positives and true negatives, ^> and <v are false negatives and false positives
the 4 numbers can be utilized to generate a large number of different calculations for classification error, prediction, performance, etc etc etc
don't memorize the whole confusion matrix – just look it up on wikipedia
accuracy of a model (true v false predictions and their overlay on reality) is not all equal!!
if you have a model that is supposed to detect something rare and isn't able to do it, the rarity of the true positive makes that model 99.9% accurate – but useless
ACCURACY = TP + TN, i.e., true predictions ÷ FP + FN + TP + TN, i.e., everything | ERROR = 1 – ACCURACY
there are many different 'scoring' performance metrics that can be generated additionally
» again, why exactly do we have this confusion matrix?
well, accuracy and error alone sometimes do not tell you enough about the model's true performance
so, we split the possible model outcomes into four different categories and conduct different nuanced scoring processes with them
rates scoring from confusion matrices
true positive rate = true positives / positives 1 – FNR
true negative rate = true negatives / negatives 1 – FPR
false positive rate = false positives / positives, or 1 – TNR
false negative rate = false negatives / negatives 1 – TPR
there is a positive-negative trade-off for each model!
moving the decision threshold cut-off for negative/positive model prediction will reduce one error at the cost of another
Type I v. Type II error [visualize two different distributions on a 3D plot]
precision/recall scoring from confusion matrices
precision [aka the positive predictive value]: true positives / predicted positives i.e., TP + FP
this tells you more about your model's ability to classify
i.e., out of the predictions you made about positive, how many did you get right
recall is just the TPR but in a precision context, i.e., there is a tradeoff between precision and recall
recall: how many positives did you get | precision: how many positives where you right about
precision y v. recall x is often graphed to describe a model
F1 score: 2 • (Pre • Rec)/(Pre + Rec), summing up their relationship
TPR = precision = sensitivity | TNR = recall = specificity
receiver operating characteristic – ROC curve
this curve describes the TPR v. FPR tradeoff as the decision threshold (i.e., your classification model) is moved around
using the ROC curve, you can determine the optimal tradeoff for your particular problem
common way to assess the performance of a model!!
AUC is area under the ROC curve, the 1-number metric that sums it up and can be used to compare models!
best AUC is 1.0 and worst AUC is 0.5 (the random guesser model)
if your AUC is less than 0.5, you have a reversed model and just flip it for better classification results!
combine ROC (AUC) and cross validation!!
generate 1 ROC curve per fold/round of cross validation
average the fold-ROC curves and take the AUC of that average ROC
this averaged AUC measures your performance and helps you handle variability when you decide on a model to use
1 note
·
View note
Text
Cross-Validation
When dataset is insufficiently big to split into training and test sets, use cross-validation
cut dataset into 5ths and assign 1/5=validation, 4/5=training in a rotating manner
The training chunks are known as a training fold and the validation set is the validation fold
with each round of train and test, use a different 1/5 of the dataset as a validation subset [without replacement after each round]
average all performances on each round of validation to get the fully-trained and tested model
subset size can be modified but too many subsets leads to too many rounds of training
to sum: cross-validation is the evaluation and averaging the performances of [smaller models] of [portions of the dataset]
instead of using an entire small dataset to train
KEY: you are not modeling your entire dataset, only portions of it
standard of splitting into subsets: 10-fold validation, k=10
LOOCV – Leave-one-out Cross Validation
the highest benchmark standard of performance that doesn't actually get utilized often because it's inefficient
in LOOCV there are N training folds
using cross-validation to select for the best model in a dataset that is large enough for a train-test split
i.e., try various models on the same data to determine which yields the best predictive models
[steps]
so split your data into train and test sets, then split the training set into those training and validation folds & vary the models and k-values used
look at the performances returned for each validation fold and select the highest-performing model
retrain the training set on that highest-performance model to get the best fit for the entire training set.
[optional: train on the whole dataset (training+testing sets) and return a whole bunch of estimates for its performance from above]
then test it on the test set!!
0 notes
Text
Regularization in ML
In linear regression, regularization counteracts overfitting
An optimizer will assign a weight to each of your datapoints in the dataset
Regularization demands that the optimizer find a weight w that falls within a certain threshold C
A higher C value decreases regularization; smaller C penalizes the slope of the line's ability to fit the dataset » slope-fit trade-off
the new function is called a regularized or augmented function
min Ein(w) + (lamba/N)w™w where as C increases, lambda decreases
watch your regularization parameter (lambda/N) – mindlessly increasing it to decrease C and regularize may result in turning the algorithm off
if the parameter is larger than the slope, the slope becomes a constant and the algorithm underfits the data
regularization normalizes the slopes of lines generated in the hypothesis space. so over-regularization over-narrows the slope distribution
knobs to control over/underfitting:
dataset size | hypothesis space | regularization parameter
modern ML is mostly the field between overfitting & regularization
regularization with gradient descent: weight decay
w(t+1) = w(t) (1-2n(lambda/N)) – n ∆ Ein(w(t))
when lambda is small, the first term goes to something less than 1 and the steps get smaller and smaller as the function goes on
0 notes
Text
Repetitive Sequences and Transposable Elements
(link to genetics notes on transposable elements here)
introduction
> 50% of the human genome is repetitive elements
simple repeats / short tandem repeats (STRs) – 3% human genome | trinucleotide repeats – common STR pattern
transposable elements – 44% human genome | eukaryotes have a larger portion of repetitive TE's than prokaryotes
large-scale duplications – 5-6% human genome | chunks of chromosome duplicated due to unequal crossing over
do these segments have a function, or are they simply either neutral or disease-causing?
expansion process = multiply repeats, generally pretty stable
i.e., short repeats can form hairpin loops on a single strand, with repeat extending after replication as a result
disease standpoint in coding regions: repeat that encodes for an amino acid may result in extra AAs and protein misfolding | EX: Huntington's
disease standpoint in noncoding regions: too many repeats results in strand/chromosomal instability & breakage and/or transcriptional suppression | EX: fragile X syndrome
sequence complexity of repeating and non-repeating segments
take the length of the sequence L and the size of the option pool (i.e., GTAC is N=4, proteins is N=20) and input them into a complexity equation
equation is K = (1/L) logN [L! / πn(i)] to find the complexity score of the sequence
you can also calculate the number of reading frame windows that could be generated from the segment, assign complexity according to a relevant threshold, and join adjacent regions w/ matching complexity
SEG and DUST complexity measures for proteins and nucleotides, respectively
transposable elements
class 1: retrotransposon – Ctrl+C, Ctrl+V
RNA copy of a DNA segment is made, moved to a different portion of the genome
RNA copy is reverse-transcribed to DNA at the new location and incorporated into the genome
LINES & SINES: see genetics notes
class 2: transposon – Ctrl+X, Ctrl+V
original DNA segment itself is excised, moves to a different portion of the genome
original DNA segment is inserted at the new location and reincorporated into the genome
autonomy of genomic movement
autonomous: all machinery is included in movement, i.e., transposase/retrotransposase genes are included within the sequence
non autonomous: relies on host proteins for movement across the genome
resolution of the C-value paradox:
genome size does not directly correlate with organismal complexity
rather the confounding variable is the individualized TE composition/replication
identifying TEs in a sequence of interest
check for repetitive flanking regions using the Dfam database, a bank of known TE sequences
RepeatMaster: identifies all occurrences of low complexity regions and specific TE repeats in a genome/region
reproductive selection for TEs
host organismal level: must be in germline without lethality or presence in host-selected for regions
genome level: need to be able to be transcribed, therefore high selection for TEs that bind host transcription factors in germline or early development
TE insertions, especially if TE binds transcription factors, can determine host gene expression. if beneficial regulatory function, the new insertion is reproductively selected for
host selection mechanisms can silence deleterious TE insertions via sRNA or DNA methylation defenses » evolutionary race
gene duplications & pseudogenes
large-scale duplications can be caused by misalignment & unequal crossing over of chromosomes
often facilitated by repetitive sequences along the genome
this leads to copy number variants in different individuals along certain portions of the genome
two-tailed beyond a certain CNV gene dosage can lead to disease and disruption of gene expression regulation
pseudogenes look like genes but don't code for proteins because they've lost some aspect of function
most often they result from duplications of a gene that were either selected to be silenced or had some inactivating mutation
another mutation in a pseudogene region could possibly reactivate it!
0 notes
Text
Conceptualizations in Machine Learning
training set error
error on training dataset, which should lower as a model is optimized
arbitrary goal is to lower the training set error
test set error
error on test dataset (reserved for testing in particular, without any contact with training process)
withholding the test set means that the test set becomes an ESTIMATE of real world data behaviors
"output"/generalization error
model's performance in the "real world" outside of training and testing datasets
different from "general gap" that defines ∆(training error, test error)
training and test set errors are estimates of this generalized error
generalizing models to data
a goal is for the model to generalize well to UNSEEN "real world" data
unfitted/untrained models are expected to have high training and test set errors
fitted/trained models are expected to have low training error and (hopefully) low test set error
underfitting
both the training and testing errors are very high because the model has insufficiently learned all the info from the data
solution: train for longer, with more data
overfitting
as more training goes on, the training error keeps decreasing and the test error suddenly starts to grow larger
essentially the model begins to "learn the noise" of the training dataset and HYPERFITS to it
the hyperfit makes the model BRITTLE with relation to testing and real-world data generalization
a simple learning problem
given: a sinusoidal function***
goal: approximate the sinusoid via your ML model
when H(0): h(x) = b, a constant, use the mean to approx. the sinusoid. this returns error across the sinusoid = 0.5
enrich the space of hypothesis! when H(0): h(x) = ax+b, the linear trendline cuts diagonally across the sinusoid. this returns error across the sinusoid = 0.2
and so on. BUT....
...in ML, you don't have the actual sinusoid*** – only a set of datapoints along that sinusoid f that give you an approx. picture of it
so training your model to a given training dataset equates to enriching/increasing the hypothesis space/complexity of the hypothesis as above
plotting training, testing, and generalized error v. hypothesis richness gives you a visualization of the underfit/overfitted model status
error v. richness for training and testing errors indicate well-fitted modeling as long as both graphs are still decreasing, even if the gap between the two grows
"sweet spot" that divides under- and overfitting occurs at the minimum of the testing error v. richness graph
going back to the constant/linear trendline hypothesis H(0): h(x) = b
taking many different hypotheses with random y-points in the dataset gives you various trendline models across the sinusoidal function
all these hypotheses average to about the same mean of the function above
so if you don't have the sinusoid f and you can't find H(0): h(x) = b from the get-go, this is how you find that average
same goes for the enriched linear hypothesis H(0): h(x) = ax + b
actually, this can only be done with a simulation in which you have multiple datasets from which to derive multiple random hypotheses that you then average. You still can't do this in the real world.
the problem of ML returns to the fact that you only have a dataset of points along some function f and not f itself. so essentially what you're doing is attempting to find the optimal H(0) for a particular richness level (constant, linear, etc) through generation of multiple random H(0)'s and averages/measures of their error
bias and variance
bias and variance are the sort-of-equivalent
low variance == precision | low bias == accuracy
the bias + variance decomposition tradeoff: Eout(x) = var(x) + bias(x)
high-variance, low-bias visualizations of a model for a function f are hard to create – imagine a 15th order polynomial made to each dataset in your data
the various 'best' hypotheses based on each dataset would look very different to one another
match the model complexity to the dataset, not the target complexity
conceptualization of the variance-bias trade-off
a mismatch between the complexity of H(0) and that of the unknown f necessarily creates some bias no matter how well your H(0) fits your dataset
on the other hand, a very complex H(0) could include f in its range, but because its range is so large, the variance is high
thus, the variance+bias trade-off
6 notes
·
View notes
Text
Epigenetics
epigenetics: genomic elements that do not affect basic nucleotide sequence of DNA itself
histone modification [explained in previous notes here]
DNA methylation at 5'C of cytosine (cytosine » 5methylatedC or 5mC) in 5'-CpG-3' and complementary 5'-GpC-3' island clump motifs
DNA methylation
methylation generally co-occurs with promoters in non-coding intronic areas to repress expression
general theory is that either a) the TF-binding site is blocked by methylation or b) methylation increases likelihood of histone incorporation
methylation in exons can either up- or downregulate expression; usually UPregulated (reasons are more poorly understood)
evolutionarily, CpG and CHG methylation percentages vary by species – and methylation effects vary by species as well
methylation identification methods
(1) digest DNA » pull down methylated regions via 5mC-specific ABs or natural 5mC-binding proteins » sequence | con: low-resolution results
(2) add sodium bisulfite to DNA, converting non-mC's to uracil » PCR to make U's to T's and non-mC-G's to A's » only 5mC's are preserved, w/ methyl group as protector
(3) the above bisulfite method to the entire genome, whole genome bisulfite sequencing (WGBS) » seq via next-gen sequencing » find % methylation at each C | con: hard to align because of C»T bisulfite alterations
(4) methylation pattern-specific PCR primers to find particular motifs and methylation sites
(5) microarrays as an alternative approach to the same endpoint of (4)
epigenetic marker propagation
i.e., does a daughter cell receive the same epigenetic markers as its parent?
DNA methylation is pretty robust during replication: the parent strand keeps its mC, and the DMNT1 protein copies methylation to the synthesized strand w/ 96% accuracy [methylation degrades over time]
histone marker propagation is less stable, even in the parent cell itself – markers stay for days only, & histones themselves are removed during replication so markers cannot be directly transferred
hypothesized histone marker propagation: a) proximity effect, where old marker goes to new histone after getting kicked off, and b) histone modification enzymes actively add same modifications to new histones during replication
generational epigenetic marker transmission
i.e., does an entire offspring receive the markers of its parent organism? » "genetic memory"
sperm and eggs have their own specific epigenetics, so markers in the adult organism's somatic cells will not transfer to the gametes
sperm themselves package DNA without???histones???
many epigenetic rewrites occur during development anyway to support tissue differentiation, so "genetic memory" would somehow need to persist through this process
HOWEVER: since developmental demethylation and loss of histone modification is NOT uniform, some parental epigenetics may remain
metabolism and trauma are two particular areas where studies yield evidence of generational epigenetic transmissions
EX: famine is transferred to F1 but not F2 from P gen | EX: POW trauma is transferred from father to son & environmentally contingent | EX: Holocaust survivor offspring have OPPOSITE effect to direct epigenetic transmission
how much of the genome is functional?
one question here is: defining "functional" in the first place!
functional: coding regions? informational v. structural? phenotypically impactful? important for survival? this definition affects considerations of % functional genome!
human genome percentages: 1.5% coding, 5% introns, < 1% functional RNAs, ~20% functional (??? purpose)
> 50% of the genome is repetitive sequences that have no informational purpose and simply self-replicate and propagate
onion paradox from reading: organismal complexity does not correlate w/ genome size
0 notes
Text
Gene Regulation II
chromatin organization
chromatin = DNA + related higher order structural packing
DNA is packed into nucleosomes with tightly-packed, non-accessible areas
use a DNAse sensitivity assay to analyze higher-order chromatin structure
chromatin organizational formatting allows for distal enhancers to be brought to promoter region for increased or decreased gene expression
cellular context
DNA occupies "chromosome territories" in the nucleus, in nuclear lamina
nuclear lamina are clustered into compartment types: accessible and inaccessible
inaccessible compartments include: lamina-associated domains (LADs) and nucleolar-associated domains (NADs)
gene expression regulation via topologically-associated domains (TADs)
a TAD includes a network of a gene, its promoter, and various enhancers that are looped together w/ cohesin proteins
an enhancer in a TAD can regulate the gene of that TAD but not other TADs' genes
Hi-C: a way to map TADs into triangular regions for analysis, that returns the level of interaction between two TADs and the genes within them using color
Hi-C: the varying color and triangular signifiers give you a boundary between two close TADs
mutations that disrupt TAD boundaries can result in an altered phenotype
new enhancers in an altered TAD region (shortened, shifted, or prolonged) have unpredictable effects on the TAD genes and result in ∆phenotype
EX: brachydactyly: branching patterns of hands diverge from the norm
EX: polydacty: more than typical number of fingers
EX: F-syndrome: fingers merge, inverse, and duplicate
histone modifications
methylation, acetylation, phosphorylation of the histone tail
code: Histone Amino_acid Position modification# i.e., H3K4me3 is "histone 3's lysine 4 is tri-methylated
[well-studied modification] active enhancer combo: H3K4me1, H3K27ac
[well-studied modification] active promoter combo: H3K4me3, H3K27ac
[well-studied modification] closed or poised (selectively active) enhancer combo: H3K4me1, H3K27me3
[well-studied modification] primed enhancer: H3K4me1
determining modification frequencies along the genome
(1) tag acetyl, methyl & other modification group linkages w/ specific ABs
(2) run ChiP-seq to find modifications and their frequencies
limitations of DNA/histone modification analyses:
correlation v. causation of modifications (i.e., a mod happens alongside a change in expression but it isn't necessarily the CAUSE of said change)
direction of elicited causations is uncertain (i.e., which one first causes the other is unclear)
ENCODE Project findings
1.5% of human genome is coding; 80% genome gets a hit for regulatory analyses at least once
so...somewhere between 1.5% and 80% of the human genome is functional
ENCODE generated a very useful MAP of modification markers and areas along the genome
0 notes
Text
Gene Regulation
prokaryote
full description found in genetics notes here
RNAP binding mechanism
operon gene cluster structure
additional notes:
regulatory region is just upstream of promoter
RNAP binds at promoter [signature binding site: -35, -10bp of gene]
positive regulation: enhancer at reg. site increases RNAP binding & transcription after small molecule binding
negative regulation: repressor at reg. site is removed by small molecule binding, which allows RNAP to bind & increase transcription
most of genome is composed of coding DNA
eukaryote
full description found in genetics notes here and here
TATA(+ WAW, where W==T or A) box
enhancer regions – activating & inhibiting
transcription factors (TFs) – activators & repressors w/ short DNA seq [seq logo] binding specificity
additional notes:
regulatory region can be up or downstream, near or far
different TFs' seq logos can be near each other in an enhancer region if they bind together
diff combos of TFs bind together for varying regulation
cohesins [also seen in meiosis] bring far-off enhancers to a promoter in a loop
insulators block certain enhancer-promoter interactions for some genes and facilitate those for others to properly regulate transcription
studying gene regulation: analytical techniques & assays
[RNA-seq]
transcriptome: set of RNA transcripts in cell
utilize RNA seq to determine changes in expression levels (think Collins lab RNA seq purposes)
∆expression levels can then tell you info about gene regulation alterations
RNA-seq general process:
gene » mRNA transcripts, so exons only » sequence RNA fragments
get double-stranded cDNA (ds cDNA) » align exons
different #s of reads generate a relative expression rate for a gene via RPKM calculations
reads per kilo base per million mapped reads RPKM = total exon reads / (mapped reads • exon length)
however! this leaves info about the actual enhancer region out, so utilize chromatin immunoprecipitation followed by deep sequencing (CHIP-seq) and chromatin interaction analysis with paired-end tag sequencing CHIA-PET
[CHIP-seq & ChIA-PET]
CHIP-seq general process:
cross-link signaling proteins to DNA of interest [enhancer region]
shear DNA into 200-600 bp fragments
add signaling protein-specific antibodies and let bind
conduct immunoprecipitation step: purify anything bound to the AB
after purification, reverse signal protein-DNA crosslink & align purified sequence to genome to locate enhancer region
ChIA-PET general process:
fix active enhancer/promoter interaction site & all bound TFs
shear the DNA loop and ligate all loose ends » sequence all DNA
find spikes in sequencing to get pol II and enhancer binding seq and genomic locations
[SELEX]
finds an unknown TF-binding motif
for a TF with an unknown binding site:
generate mutant library of potential binding sites
select all fragments that are able to bind the TF
sequence those fragments & conduct PCR to add them again to the library
thus, find the motif that the TF binds to
0 notes